Single nucleus RNA sequencing is an alternative version of single-cell RNA sequencing with the 10x Genomics Single Cell Gene Expression solution. As the name suggests, sequencing is preceded by a nucleus isolation step instead of tissue dissociation into single cells. When is this alternative suitable?
Single nucleus RNA sequencing — Not all sample types are suited for single-cell dissociation, as the cell is difficult to separate from its matrix without disrupting its valuable RNA content.
Here, the extracted nucleus, protected by its bilipid envelope, confers a lower chance of rupture and its RNA contents are more likely to retain their natural composition. Hence, performing nucleus isolation instead of tissue dissociation – a technique called single nucleus RNA sequencing – is an outcome, serving as a preferred alternative to whole-cell transcriptomics.
10x Genomics Nuclei Isolation Protocol
This is true in some cases, but not all. When is single nucleus RNA sequencing relevant? How can you know if the tissue you’re working on benefits from discarding the cytosol and continuing with the core? And what limitations does the technology have?
If required, Single Cell Discoveries offers nucleus isolation on your submitted tissue. In other words, we take over the entire process of single nucleus RNA sequencing, including isolation, sample preparation, sequencing, and exploratory data analysis steps. As always, we provide a complete solution.
Click here to jump to a specific section:
- Advantages
- Tissues that require single nucleus RNA sequencing
- Limitations
- Which tissues are incompatible with nucleus isolation and why?
- Alternatives
- Conclusion
Advantages of single nucleus RNA sequencing
What are the benefits of single nucleus RNA sequencing?
- Intact single-cell dissociation can be difficult for some tissues, such as brain, heart, adipose tissue, or skin. For these tissue types, nuclei isolation is often an easier protocol than single-cell dissociation.
- Single nucleus RNA sequencing circumvents at least two types of biases:
- Cell population biases caused by cell dissociation. Such biases have sometimes been observed in the human kidney and other tissues (Van den Brink et al., 2017).
- Cell capture bias caused by variations in cell size (in a microfluidics system).
- Nuclei isolation causes less cell stress than single-cell dissociation. This prevents the occurrence of stress-induced transcriptional changes (artifacts).
- Performance is found to be comparable to single cell RNA sequencing in most cases, although this may differ per sample type and comparisons for some tissues are lacking.
- Nuclei isolation is a favored outcome for sequencing frozen whole tissue, on which dissociation can lead to low viability. However, 10x Flex, launched in 2021, is the current preferred method for sequencing frozen human and mouse tissue.
- To attain single-cell chromatin accessibility profiles with Single Cell ATAC or Multiome, nuclei isolation is a required step.
- Studies focusing on nuclear RNAs or studies using nascent (immature) mRNA levels to reconstruct gene expression changes over time, may have more success with single nucleus RNA sequencing.
Which tissues are incompatible with dissociation and why?
Several tissues are notoriously difficult to dissociate, as many neuroscientists and cardiovascular researchers are well-aware. What causes this? And for which tissues is single nucleus RNA sequencing a solution?
Below we document a collection of tissue types for which we know single nucleus RNA sequencing is or has often been a solution. Know that this is not an exhaustive list due to the scale of possible tissue types.
Central nervous system
Brain and spinal cord tissue are notoriously hard to dissociate intact. Partly, this is because large amounts of fatty myelin interrupts lysis. And partly, this is because large-sized neurons’ axons or dendrites are difficult to prepare. The technical challenges of preparing single-cell suspensions have thus prompted researchers to test other techniques.
From the start of the single cell revolution, single nucleus RNA sequencing has been the golden standard for central nervous system tissue. However, the relatively recent launch of 10x Flex, which is compatible with frozen tissue by a formaldehyde-fixation step (more info below), is changing this.
Heart tissue
Similar to brain dissociation, proper sample preparation of heart tissue is technically challenging and requires a combination of enzymatic and physical dissociation. Moreover, the high-throughput 10x Genomics system restricts cell size to around 30 µm, while cardiomyocytes (as many muscle cells) are usually larger. Hence, though strictly speaking dissociation is possible, single nucleus RNA sequencing is often the preferred option (Lacar et al., 2016).
Importantly, whole-cell cardiomyocyte sequencing is possible with SORT-seq, a plate-based single-cell sequencing technology exclusively offered by Single Cell Discoveries. This may be a suitable outcome if high-throughput sequencing is not required.
Tumor and precancerous tissue
Most cancer tissue is compatible with both cellular dissociation and nucleus isolation. Since patient-derived tumor tissue is also commonly frozen immediately after resection surgery, many researchers performed single-nucleus RNA sequencing until 10x Flex became a possibility (more information on 10x Flex below). There is still at least one exception.
Research into precancerous tissue such as fibrotic tissue can benefit from single nuclei sequencing. To give just two examples, in fibrotic liver and kidney, cell dissociation can result in a bias toward certain cell types. In fibrotic liver, immune cells seem to be captured more effectively than hepatic and tumor cells (Wen et al., 2022). In fibrotic kidneys, cell dissociations may miss most glomerular podocytes, mesangial cells, and endothelial cells (Haojia et al., 2019).
Get the 10x Genomics information guide
For a comprehensive summary of our 10x Genomics services, including single nucleus sequencing, 10x Flex, and more, you can find our information guide.
Non-pharmaceutical model species, insects, and plants
Single nucleus RNA sequencing is a preferred method for frozen tissue of non-human or -mouse origin (see 10x Flex discussed at Alternatives for the reason why). Frozen tissue is omnipresent in research as it achieves high-quality preservation for storage and transport, so this option is common throughout research areas. Examples include frozen zebrafish, salmon, sheep, chicken, rabbit, nurse sharks, and pig.
In mature (non-larvae) insects, such as the common experimental model Drosophila (fruit fly), cell dissociation is impaired due to their exoskeleton. Plants, Arabidopsis or rice to give two common study objects, have cellulose cell walls which provide a similar obstacle.
This does not mean single nucleus or cell sequencing is impossible. Though not straightforward, both Drosophila nuclei isolation and cell dissociation protocols exist (Li et al., 2022, McLaughlin et al., 2022). One may be preferred over the other depending on context. For plants, enzymatic cell wall digestion and manual isolation are the two methods for single cell isolation. Both, however, require custom optimization and verification per plant species. The development of nucleus isolation provides an outcome for some in which dissociation fails (Tian et al., 2020).
We have experience with several plant and insect species — feel free to contact us for details.
Your species not on this list?
Contact us to get the most relevant information and advice that we have for your specific technical or biological question.
Sequenced Species at Single Cell Discoveries

Limitations of single nucleus RNA sequencing
What are the limitations of single nucleus RNA sequencing?
- Nuclei have lower mRNA levels than whole cells, so the total number of detected genes and UMIs per cell is generally less. This also means that, during data analysis, cutoffs that usually account for cell quality like genes per cell, UMIs per cell, and mitochondrial genes per cell, are lowered. Theoretically, this increases the chances of accepting perturbed cells in the analysis. However, effects on data quality are probably minimal. Ultimately, sensitivity differences between single nucleus and single cell RNA sequencing depend largely on the sample type.
- Single nucleus RNA sequencing requires additional reagents, additional steps, and additional personnel compared to single-cell RNA sequencing.
- Nuclear pores are accessible to disruptive RNAse enzymes, so the protocol requires working with RNAse inhibitors and all steps should take place in a cold environment. This cool milieu also decreases chances of artefactual transcriptional changes.
- Single nuclei RNA sequencing often requires more tissue to compensate for loss of material during nucleus isolation. At least 60 mg of tissue is a common sample requirement, compared to, e.g., 12 mg for 10x Flex or 20-50 mg for dissociated cells.
- Not all tissues invariably show good results of nucleus isolation (see next chapter).
- The flip side of a bias toward nuclear RNA is, logically, the loss of cytoplasmic RNAs (mRNAs, ribosomal RNAs, mitochondrial RNAs). In theory, this makes the attained single-cell gene expression profile less complete and accurate.
- To summarize, for an easily dissociated tissue, the “costs” of nuclei isolation generally outweigh the benefits.
Which tissues are incompatible with nucleus isolation and why?
10x Genomics documents a lot of in-house testing of nucleus isolation of a wide variety of human and mouse tissue types. This includes healthy tissue like mouse brain, eye, and colon and human jejunum, ileum, and testis, as well as diseased tissue like the most frequent tumors (breast, prostate, lung among others).
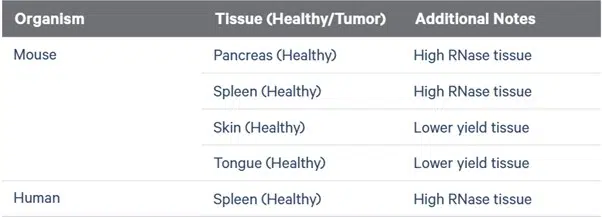
However, some tissues show variable results and may require further optimization:
Source: 10x Genomics nuclei isolation user guide.
Other tissues are deemed incompatible with the 10x Genomics protocol, although alternative methods exist. These are:
- As mentioned before, the plant cell wall presents a significant barrier to cell dissociation. In addition, plant nuclei differ from animal nuclei in size and debris so much, that they require specialized protocols. These protocols, however, do exist.
- Mature insects and similar species. The insect exoskeleton prevents intact dissociation and chitin-containing tissues interfere with the lysis step for nuclei isolation. This group also includes some fungi, lobsters, and arachnids. As mentioned, single nuclei sequencing protocols for Drosophila (fruit fly) do, however, exist (e.g., McLaughlin et al., 2022. Li et al., 2022).
- Bone and other calcified tissues. The lysis step in nuclei isolation is often unsuccessful in bone or other calcified tissue. However, single-cell RNA sequencing is done in orthopedic research (see Wang et al., 2023).
- FFPE tissue. See the discussion of 10x Flex in the chapter on alternatives to single nucleus RNA sequencing.
In addition, the requirement of high tissue volume can exclude some tissues from single-nucleus RNA sequencing. Young mouse or guinea pig brains, as well as specific embryonic structures in humans or other species, may be too small to reach the requirements.
You can always contact us if you are curious about our experience with the tissues mentioned above.
Alternatives to single nucleus RNA sequencing
Sometimes, neither single nucleus RNA sequencing nor single-cell dissociation is an option. Perhaps no nuclei isolation protocol exists for a specific tissue. You may make a specific consideration of costs and resources that makes you search for alternatives. Or the limitations mentioned above obstruct the successful achievement of your biological question.
Fortunately, there are still alternatives to single nucleus RNA sequencing even when single-cell dissociation is a priori unpracticable.
Optimize sample purification
Get rid of obstructive material by, e.g., sorting or centrifugation, and gradually increase the quality of cells by protocol optimization. You can leverage the broad experience and single-cell quality control expertise of Single Cell Discoveries’ scientists and research and development unit. We also perform pilots to explore possibilities without extravagant waste of tissue or resources.
10x Genomics Single Cell Gene Expression Flex
A shift in preferred technology has occurred over the past two years, especially for neuroscientists and researchers working with frozen patient or mouse material, since the availability of 10x Genomics Single Cell Gene Expression Flex (10x Flex).
For reasons discussed above, single nucleus RNA sequencing achieved higher-quality results on frozen tissue than single-cell sequencing. However, 10x Flex now seems to outperform single nucleus RNA sequencing and is the advised technology for most non-fresh human and mouse tissue types.
In the protocol, frozen, FFPE, or OCT embedded tissues are thawed and formaldehyde-fixed, then deparaffinized or pulverized instead of dissociated. This retains RNA quality better than thawing and single-cell dissociation, requires up to five times less starting material than single nucleus RNA sequencing, and has other benefits (and limitations) too.
Why only human and mouse? 10x Flex differs from other protocols mainly in the fact that it uses human- or mouse-specific mRNA-targeting probes to capture the mRNA of single cells. Although theoretically possible, utilizing these probes on other species results in significantly low sensitivity, making other techniques preferable.
For pig heart tissue (common in cardiovascular research) or non-human primate brain tissue (common in preclinical testing of neurological drugs), single nucleus RNA sequencing is still often most suitable.
Human-mouse xenograft tissue (i.e. patient- or cell line-derived xenografts, PDx resp. CDx) is possible too, an application recently developed by our research and development team.
For more information on 10x Flex: click here
For more information on 10x Flex for PDx or CDx tissue: click here
Conclusions
In almost all cases, the choice for single nucleus RNA sequencing over single cell RNA sequencing depends on the context: the exact tissue types, technical details of the available tissue, the biological question, and the available resources.
It is wise to check the scientific literature on single-cell sequencing of your specific tissue, although it is important to consider that best practices might change.
Don’t hesitate to reach out if you are curious whether single nucleus or single cell RNA sequencing is the advised technique on a specific sample. Want to know the most up-to-date advice on tissues mentioned in this blog post? Our team is always happy to assist.
Find out more
Find out more information on single nucleus isolation, our other 10x Genomics services, inspiring resources, and much more in our information guide: