CITE-seq is a method to simultaneously perform single-cell RNA sequencing and single-cell surface protein analysis. But how does this work? And what does it accomplish?
Single-cell RNA sequencing (often abbreviated to scRNA-seq) is a powerful technology that allows researchers to measure the gene expression of individual cells. This has provided valuable insights into how gene expression varies across different cells in a tissue and has contributed to our understanding of many biological processes.
CITE-seq has been developed to allow simultaneous measurement of individual cells' gene expression and the proteins they express on the cell surface. Because cell-surface proteins are often useful markers for cell identity or cell state, this compatible technology can expand the power of single-cell analysis.
In this blog, we will discuss the benefits of this technology and how researchers use it. Here are the topics we cover:
- What is CITE-seq?
- What does CITE-seq stand for?
- How does it work?
- TotalSeq: how Single Cell Discoveries performs CITE-seq.
- What are its advantages?
- What are its limitations?
- How is it applied?
What is CITE-seq?
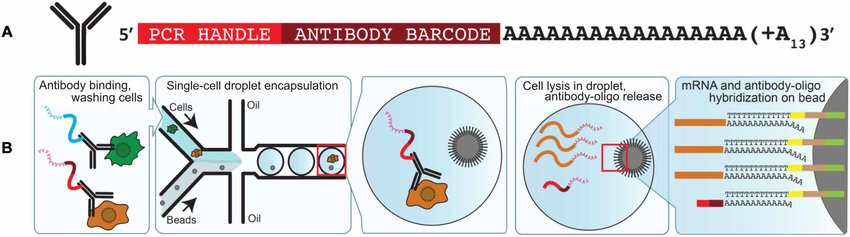
CITE-seq is a method of labeling cell-surface proteins with uniquely barcoded antibodies, which can then be captured and quantified along with the RNA molecules in single-cell RNA sequencing. As a result, CITE-seq data contains information on the transcriptome and cell surface proteins.
What does CITE-seq stand for?
CITE-seq stands for Cellular Indexing of Transcriptomes and Epitopes by Sequencing. You can understand the term as follows:
- Cellular Indexing in this context means adding a cell-specific barcode to both the RNA fragments and cell surface proteins;
- Transcriptomes are one of the indexed datasets; it is the gene expression data, also known as the RNA profile;
- Epitopes in this context mean the cell-surface proteins, named as such because antibodies recognize them —this is the second dataset;
- Sequencing is the detection method, i.e., the reading of the DNA barcodes that identify transcript and epitope.
How does CITE-seq work, exactly?
CITE-seq combines the process of single-cell RNA sequencing with cell-surface protein detection. We will explain both technologies.
Single-cell RNA sequencing
CITE-seq is compatible with 10x Genomics technology. This high-throughput single-cell RNA sequencing technology uses a microfluidic system based on an oil-water emulsion. How does this work?
Get the 10x Genomics information guide
For a visual explanation of 10x Genomics, CITE-seq, cell technologies you can find our information guide.
First, tissues need to be dissociated into a single-cell suspension by a tissue-specific protocol. Then, inside a Chromium X or comparable machine, the cells are partitioned into water droplets with reagents and barcoded gel beads. These barcoded gel beads are covered with oligonucleotides: short stretches of nucleotides.
Each oligo contains two barcoding elements and one RNA-binding element. This latter part is a poly(T)-stretch which binds the mRNA. One barcoding element is a unique barcode for each bead. This is tagged onto the mRNA and makes it traceable to the cell it came from. Another part of the oligo uniquely barcodes each single RNA molecule. It is called the unique molecular identifier, a.k.a. UMI, and is important to quantify gene expression accurately.
The filled water-droplets-in-oil are called GEMs, and each GEM should contain one cell, one barcoded gel bead, and reagents. In each GEM, the reagents cause the cells to release the RNA, and the mRNA molecules bind to the barcoded gel beads. Then, the GEMs are collected, and a process of several steps ultimately creates copies of barcoded DNA fragments.
Each fragment includes both unique barcodes from the gel bead, plus a part of the mRNA code, so each fragment contains all information necessary to link an mRNA molecule to its specific gene and cell. After next-generation sequencing and data analysis, you've established a transcriptomic profile of the single cells in the tissue.
Cell-surface protein detection
The protein detection part is based on antibody technology. CITE-seq uses antibodies specifically manufactured to bind to a cell-surface protein of interest. Say that the protein of interest is CD4, a surface marker for T-helper cells. You can add anti-CD4 antibodies to the cell mix so they will bind to all cells that express CD4. Then, you wash all unbound antibodies away.
What makes the two technologies compatible
The antibodies are connected to a unique oligo compatible with the 10x Genomics platform. In our example, the oligo contains a barcode specific to the anti-CD4 antibodies. This is necessary to trace the protein back to the cell it was on. The oligo also includes a poly(A)-stretch that can bind to the gel beads. This component allows for the simultaneous detection of protein and RNA in one experiment.
Because the antibodies attach to the individual cells, they end up together in one GEM. Here, they get tagged with the cellular barcode and unique molecular identifier from the gel beads. The oligos separate at the right moment and are captured and quantified along with the mRNA molecules.
In the sequencing data, the specific CITE-seq barcodes, also called antibody-derived tags (ADT), identify the antibody bound to the cell and infer the cell-surface protein present. In our example, this makes it possible to quantify the CD4 molecules of each cell.
Simultaneous quantification of multiple proteins
You can combine multiple different antibodies in one experiment. After adding a cocktail of different barcoded antibodies to a sample, the antibodies can bind to their corresponding epitopes. All unbound antibodies are washed away to not interfere with the technology. This is often necessary in immunology, where two or more surface proteins may characterize cell subtypes and their activation states.
How do we perform CITE-seq?
Single Cell Discoveries uses a version of CITE-seq called TotalSeq, which uses antibody-oligonucleotide conjugates from BioLegend, a global antibody manufacturer.
We can combine TotalSeq with the 10x Genomics Single Cell Gene Expression solution and Single Cell Immune Profiling solution.
The Single Cell Gene Expression solution provides users with 3’ RNA expression at single-cell resolution. This high-throughput sequencing solution is highly suitable for larger projects that require data for a few thousand cells per sample.
The Single Cell Immune Profiling solution provides users with 5’ transcriptome analysis combined with the T- and B-cell repertoire and antigen specificity. The objective is to sequence the 5' end of mRNA molecules because, in T and B cells, this stretch contains the variable region of the T- and B-cell receptor that makes the receptor recognize specific antigens. With this solution, you can explore immune cell diversity or characterize the immune response to infection or treatment.
Download the 10x Genomics Information Guide
TotalSeq-B vs. TotalSeq-C
Both solutions require antibodies in which the oligos are slightly different. We combine TotalSeq-B with the 10x Genomics Single Cell Gene Expression solution and TotalSeq-C with the 10x Genomics Single Cell Immune Profiling solution.
TotalSeq-B antibody oligos can attach to the oligos present on the Single Cell Gene Expression gel beads. In contrast to CITE-seq, they adhere to separate oligos from the ones to which RNA molecules attach (see figure). The antibodies bind to a nucleotide stretch on these oligos called CaptureSeq1.
TotalSeq-C antibody oligos can attach to the Single Cell Immune Profiling gel beads. Here, the 5' end of the RNA molecules and the antibody oligos attach to the same gel bead oligo (see figure). They both bind to a nucleotide stretch on these oligos called TSO.
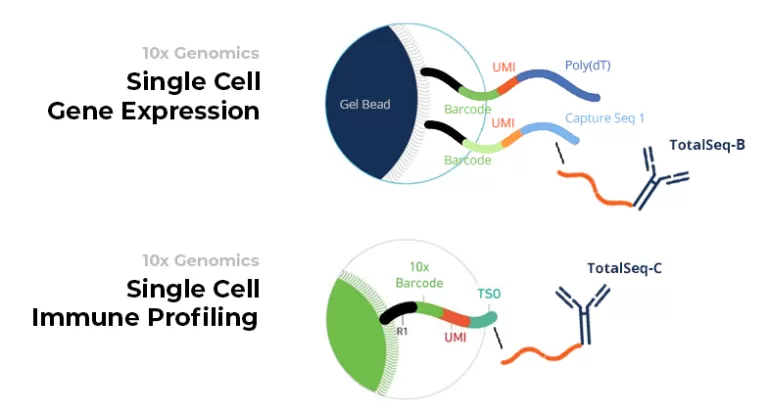
Credits: 10x Genomics 3' and 5' protocol, and BioLegend.
Antibody cocktails
We can provide one of two antibody mixes for both solutions.
- The human TBNK cocktail. This mix contains antibodies specific for the nine surface proteins CD19, CD3, CD16, CD4, CD11c, CD56 (NCAM), CD14, CD8, and CD45. With this cocktail, you can characterize immune cell subtypes such as T cell subsets, B cells, natural killer cells, antigen-presenting cells, and dendritic cells.
- The Human Universal Cocktail. This mix contains antibodies specific to 130–134 different epitopes (find out which for TotalSeq-B or TotalSeq-C). It helps characterize major immune cell lineages and selected cell states.
What are the advantages of CITE-seq?
Combining single-cell RNA sequencing with cell surface protein analysis provides a comprehensive approach.
CITE-seq, for instance, directly links a cell's RNA profile with its cell-surface protein expression, offering a significant advantage. This combination is crucial because proteins are the central functional units of a cell and play a vital role in various cellular processes. Sometimes, a cell population may have abundant surface proteins in only a fraction of the cells, as noted by Peterson et al. (2017). To address this discrepancy, researchers can merge both measurements.
In certain instances, distinguishing cell subsets based on their RNA profiles alone can be challenging. Multimodal data, such as transcriptome plus cell-surface proteins, aids in identifying subsets that might otherwise be missed. For example, in a proof-of-concept study by Stoeckius et al. (2017), researchers successfully identified natural killer cell subsets that were challenging to differentiate based solely on RNA-seq data. More recently, Ben-Chetrit et al. (2023) applied a novel spatial variant of CITE-seq to a mouse breast tumor, demonstrating their ability to distinguish between two cell states using protein markers: tumor-fighting and immune-suppressive macrophages.
CITE-seq, by simultaneously analyzing transcriptome and cell surface proteome, enables researchers to explore the interaction between these two layers of information at the single-cell level. This holistic approach provides a more profound understanding of biological processes within cell populations during health or disease.
This integrated approach is particularly valuable for pharmaceutical applications, given that many cell-surface proteins serve as diagnostic biomarkers or therapeutic targets. In fact, approximately two-thirds of the targets for approved human drugs listed in the DrugBank database are surface proteins (see also this paper). This highlights the importance of approaches like CITE-seq in drug development.
What are the limitations of CITE-seq?
As with any technology, CITE-seq has certain technical limitations.
CITE-seq and TotalSeq are limited to tagging surface proteins. Access to epitopes inside a cell is not possible because it would require puncturing the cell surface membrane. That would release the cell’s RNA content, making the sample incompatible with current RNA sequencing technology.
There is no barcoded antibody for every surface protein, so the application of CITE-seq is limited to the manufactured antibodies available. For example, there are more barcoded antibodies for human or mouse epitopes than for other species. Moreover, measuring surface proteins outside of those targeted by universal antibody cocktails will be inaccessible for some researchers because labeling antibodies can become very expensive. New antibodies and antibody cocktails will become accessible as the field develops.
Surface proteins may change during experimental steps. For example, dissociating cells with enzymes that digest proteins may also affect surface protein analysis. Also, an antibody’s binding to a surface protein could activate a cell’s signaling pathway and thereby change the cell’s transcriptome. So, experienced users recommend fixing cells on ice and testing the effect of dissociation and antibody binding on a specific tissue.
The reliability of CITE-seq results may depend on the specificity with which antibodies bind to an epitope. There’s also an increased risk of cross-reactivity in mixes of multiple antibodies.
An alternative approach for simultaneous cell-surface protein analysis with single-cell RNA sequencing is combining flow cytometry (FACS) with plate-based sequencing methods like SORT-seq or VASA-seq. When should you choose this method over CITE-seq? Generally, what you lose in volume and speed, you gain in accuracy and modal flexibility. But if you’re interested in performing simultaneous transcriptome and surface protein analysis on a specific sample, you should always reach out for a consultation.
How researchers apply CITE-seq
One of the key applications of CITE-seq is studying immune cells. The immune system comprises a diverse array of cell types, each of which plays a unique role in the body's defense against infection and disease.
Using CITE-seq, researchers can measure individual immune cells' gene and protein expression, providing valuable insights into how these cells function and respond to different stimuli.
Decades of research with fluorescent antibodies and flow cytometry assays have created an extensive knowledge base linking cell-surface proteins with immune cell type and function. This knowledge can be employed with single-cell RNA sequencing to better identify subsets and activation states of immune cells in a tissue. For example, a group of scientists from Amsterdam UMC used CITE-seq to study which immune cells reacted to SARS-CoV-2 in people unexposed to either coronavirus or -vaccine (read about it here).

High-throughput sequencing makes it possible to analyze large groups of (immune) cells all at once. For example, one group of scientists performed CITE-seq on an entire atherosclerotic plaque to study T-cell heterogeneity. This enables the study of the immune system in greater detail and can help researchers develop better therapies for treating immunological diseases and improving the body's immune response.
Another area where CITE-seq is being used is in the study of cancer. Tumors are heterogeneous tissue that pools of different immune cells often infiltrate. Multimodal information on cancer and immune cells can improve our understanding of how the tumor immune microenvironment progresses and help researchers develop more effective cancer treatments.
Concluding remarks
In all, you can view CITE-seq (and, equally, TotalSeq) as a valuable expansion of single-cell RNA sequencing. As a researcher, it allows you to combine decades of knowledge on cell-surface protein analysis with exceedingly complete single-cell transcriptome data. It is applied in immunology, oncology, and other fields for fundamental and translational questions.
Further reading
Find out more information on fixed RNA profiling, our other 10x Genomics services, resources, and much more in our information guide: