
10x Genomics Multiome - 10x Genomics' combinatory technology Multiome Single Cell Gene Expression + ATAC can be a perfect tool for many intricate questions in drug development.
In two blog posts, we aim to help researchers determine when and why to apply the technology. How exactly does it work? What questions is 10x Multiome particularly powerful to answer? And how does 10x Multiome compare to standalone scRNA-seq or scATAC-seq?
Find our first blog here: How 10x Multiome Works: A Guide
Below, we delve into nine prime questions resolvable with 10x Multiome. This section divides into 10x Multiome’s capacity to (1) deeply characterize cell populations, and (2) map regulatory networks at single-cell resolution. Thereafter, we compare 10x Multiome's benefits and limitations to its standalone counterpart technologies.
Our clients observe that the fastest way to find out if this technology suits your particular case is to contact our consultants.
Deep characterization of cell populations
1. Which cell populations exist?
Chromatin accessibility and gene expression are mechanistically connected. Chromatin accessibility defines the access of transcription factors and chromatin remodelers to DNA, which is highly associated with cis-regulatory actions in the genome and underlies cell-type-specific gene expression.
With 10x Multiome, you can access the combinatorial power to identify cell types in a tissue by grouping together nuclei with similar gene expression and chromatin accessibility profiles. Hence, the combined data serves for cross-validation, creating a more in-depth picture of cell populations and doubling the layers of information with which to annotate a cell’s type or state.
2. Are there “primed” cells?
scRNA-seq and scATAC-seq capture a cell’s state at a single point in time. With 10x Multiome, it is also possible to gauge the state towards which a cell is progressing.
Cells may be in one state, observable in their gene expression profile, but already show the gene expression they are preparing for in their chromatin accessibility profile. In part, this is due to a natural delay between transcription initiation and finalized transcription. In addition, researchers have observed that cells may prepare their chromatin accessibility further in advance, “priming” themselves for a gene expression shift like a cocked gun (Ma et al., 2020).
In addition, it is possible to identify transcription factors and regulatory mechanisms responsible for priming cells, as explained in more detail in another section.
This makes 10x Multiome highly suitable for high-resolution mapping of cell fates in developmental biology and stem cell research.
3. Are there cells marked by unique chromatin-plus-expression profiles?
In addition to revealing “primed” cells, 10x Multiome can unearth novel cell types that are undistinguished by gene expression or chromatin accessibility alone, yet show a unique combination of gene expression and chromatin accessibility profile deviant from other cell types. Examples include transitioning intermediates or stem cell–like subpopulations with regenerative potential (Bi et al., 2021)
Let's dive into an example of how 10x Multiome answers this type of question.
Matched scRNA-seq with scATAC-seq data per nucleus
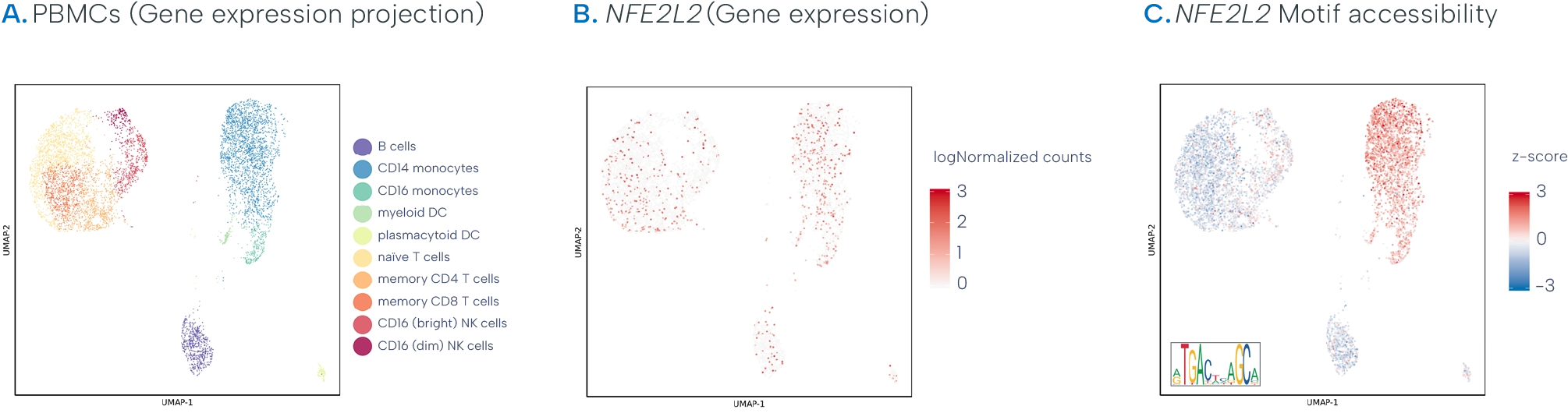
10x Genomics Multiome example data - Each dot represents a nucleus extracted from healthy peripheral blood mononuclear cells (PBMCs), processed with 10x Multiome. Source: 10x Genomics
A. Cluster analysis results from 7,273 nuclei using gene expression data. Cell populations are manually annotated based on established marker genes.
B. Zooming in, we observe expression differences of the transcription factor NFE2L2 across cell types.
C. However, we see that the NFE2L2 motif (discussed more below) accessibility, derived from ATAC data from the same cells, is specific to monocyte populations. The difference in NFE2L2 expression and motif accessibility is likely a reflection of its functional status. Normally, protein translated from NFE2L2 remains sequestered in the cytoplasm but, in response to oxidative stress, will translocate to the nucleus to regulate the expression of antioxidant proteins.
4. How do chromatin accessibility and gene expression change over time?
10x Multiome applied to time-course experiments provides a dataset highly suitable for tracing cell populations over time. In particular, “primed” cells at an earlier time point may be convincingly linked to differentiated cells at a later time point. As can consecutive cell states with known epigenetic and transcriptional markers. The resulting roadmaps are often strengthened by findings relating to the regulatory networks of cells, described fully in the next section.
The technology is often applied to generating high-resolution roadmaps of cell fates in samples including:
- Developing tissues (e.g., Ma et al., 2020; Frazel et al. 2023)
- Stem cell-based model systems such as organoids (Lee et al., 2023)
- Immune cell lineages (Chopp et al., 2023)
- Transition processes in cancer (Han et al., 2022)
Get the 10x Genomics
information guide
You can find a full explanation of 10x Genomics, its variations, and our approach in our information guide.
Map regulatory networks in single cells
5. What regulatory elements are active?
Probably the most common utility of 10x Multiome is to enhance a sample’s single-cell transcription profile with an additional layer of information about each cell’s regulatory elements.
The organization of accessible chromatin across the genome namely reflects a cell’s network of possible physical interactions through which enhancers, promoters, insulators, and transcription factors regulate gene expression. Accessible chromatin at the location of a regulatory element (a “peak” in the scATAC sequencing library) confers that this regulatory element is likely active.
6. Where are transcription factors binding?
Another common aim of chromatin accessibility analysis is to identify known and novel sequence patterns occurring within peak regions. These can be binding motifs, short sequences of DNA to which transcription factors bind to regulate gene expression.
10x Multiome connects three layers of information:
- Expressed transcription factors in the gene expression profile.
- Binding motifs of transcription factors and regulatory element activity in the open chromatin profile.
- The products of activated gene expression in the gene expression profile. This enables the generation of refined data that can improve both the accuracy and success rate of motif discovery.
10x Genomics' Multiome three layers of connected information

7. Which regulatory elements drive gene expression, how?
In a further step, the chromatin accessibility profile produced by 10x Multiome links active regulatory elements directly with gene expression. Indeed, 10x Multiome’s ability to detect both the gene expression regulator and its product (mRNA transcript) is a relatively confident indication of which regulatory elements are active in individual cells of a tissue.
This allows you to reconstruct causal connections between regulatory elements and gene expression. At a larger scale, this enables researchers to model tissue development, dissect immune cell reactivity, or identify specific regulatory elements or programs that drive disease.
An example is the application of 10x Multiome to reveal the mechanism by which chromatin remodeling complex protein PBAF promotes CD8+ T cell differentiation in response to viral infection and cancer (Kharel et al., 2023).
8. What are the unique gene networks in each cell type?
Answers to the three questions above combine to shape a high-resolution, single-cell map of gene and regulatory networks in a tissue. This gene network map can characterize cell types uniquely present in a tissue, in disease, or during a tissue’s transition phase.
For example, a group of researchers applied 10x Multiome to 11 different cancer types to compile a pan-cancer list of epigenetic programs involved in metastasis (Terekhanova et al., 2023).
9. How do conditions influence regulatory elements and downstream pathways?
Finally, comparing gene expression and chromatin accessibility changes of tissues subjected to different experimental conditions, genetic manipulation, or therapeutics, can reveal a high-resolution view of tissue response.
For instance, you can unearth the mechanism of action of a therapy that is expected to initiate a complex, heterogeneous response, a common effect of immuno-oncology, gene therapy, and cell therapy platforms.
An example is 10x Multiome applied to identify mechanisms of resistance in multiple myeloma cancer patients who underwent monoclonal antibody therapy in a clinical trial (Derrien et al., 2023). With the technology, the team implicated a genetic inactivation as well as an epigenetic silencing of regulatory elements to underlie resistance.
10x Multiome vs. standalone experiments
10x Genomics' Multiome combines two techniques to enable data capture on two levels from a single cell. Hence, it might be interesting for researchers who are considering or have experience with one of the technologies, to incorporate the other. For these people, it is important to know the limits of 10x Multiome powers. How does the combinatory technology compare to its standalone parts?
10x Multiome quality versus standalone snRNA-seq
The quality of the gene expression profile captured in 10x Multiome is ostensibly comparable to that of standalone single-nucleus RNA sequencing (snRNA-seq). 10x Genomics reports on in-house comparisons showing sensitivity, as measured by median genes and UMIs per nucleus, is only slightly lower for 10x Multiome (technical note, 10x Genomics). The report shows no effect on cell clustering, cell type proportions, and cell markers.
However, it can depend on the tissue type whether Multiome is producing similar quality gene expression results as scRNA-seq. Contact our consultants to learn about our experience with your tissue type.

scRNA-seq quality comparison by genes per cell and UMIs per cell, between 10x Multiome and standalone 3' Gene Expression. Source: 10x Genomics
Mandatory nuclei isolation
Additionally, nuclei isolation is mandatory for 10x Multiome because it is a requisite for scATAC-seq’s tagmentation step. This contrasts with scRNA-seq, which can be performed on nuclei and whole cells. You can get an idea of how important the whole-cell transcriptome would be for your experiment in our informative blog on single-nucleus RNA sequencing.
A workaround is to combine a standalone whole-cell scRNA-seq experiment with a standalone (single-nuclei) ATAC-seq experiment by dividing the sample for two separate analyses.
10x Multiome versus standalone scATAC-seq
Compared to standalone scATAC-seq, 10x Multiome is currently outperformed in terms of sensitivity and library complexity. In a systematic benchmark study on peripheral blood mononuclear cells (De Rop et al., 2023), 10x Multiome produced half the unique fragment peaks as the most advanced 10x Single Cell ATAC protocol.
The study reports that 10x Multiome results in additional costs while it is less sensitive and efficient in sequencing that standalone scATAC-seq. This has to be taken into account in designs for which scATAC-seq is the primary focus of a study. For these designs, 10x Genomics Single Cell ATAC may be the preferred option.
How to access 10x Multiome
At Single Cell Discoveries, we recommend discussing your plans with one of our consultants. This will help you decide whether 10x Multiome is the most suitable technology for your purpose. Perhaps a standalone scRNA-seq or scATAC-seq experiment will be the most successful option. Our tailor-made approach also makes it possible for us to offer customized protocols and new assays.
We work with pilots that enable you to try out a preferred approach before committing to a larger-scale project. Moreover, we have sequencing and data analysis in-house that you can leverage to answer the biological question of your research completely.
More information
Find more information on 10x Genomics, and Multiome, and our approach in our information guide.
Get the 10x Genomics information guide




