10x Multiome - Almost all medical diseases in need of new therapeutics—including cancer, immune diseases, and neurodegenerative disorders—result from a complex interplay between gene expression and epigenetic programs.
In order to understand the pathological processes underlying these diseases, and find effective treatments for them, many researchers thus turn to multimodal single-cell analysis.
The dominant technology in this field is 10x Genomics’ Single Cell Multiome ATAC + Gene Expression solution (10x Multiome). It integrates single-cell RNA sequencing (scRNA-seq) with a single-cell epigenetic readout in the form of chromatin accessibility profiling with single-cell ATAC-seq (scATAC-seq).
10x Multiome can be the most suitable tool for many intricate questions in biology and medicine. In two blog posts, we aim to help researchers determine when and why to apply the technology. How exactly does it work? What questions is 10x Multiome particularly powerful to answer? And what are the pros and cons of 10x Multiome compared to standalone scRNA-seq or scATAC-seq?
Our clients observe that the fastest way to find out if this technology suits your particular case is to contact our consultants.
How 10x Multiome works in a nutshell
10x Multiome is a high-throughput single-cell method that aims to capture the transcriptome and chromatin accessibility profiles of individual cells. Its technology combines a pre-existing scRNA-seq with a scATAC-seq technology, namely 10x Genomics’ (3’) Chromium Single Cell Gene Expression and Chromium Single Cell ATAC.
3’ Chromium Single Cell Gene Expression
Single Cell Gene Expression captures the gene expression profile of all single cells in a tissue by capturing and sequencing the mRNA transcripts in a cell. That allows you to assess the state of single cells, perform cell type identification, and much more.
The protocol’s first step is single-cell dissociation or nuclei isolation. This is followed by a single-cell barcoding step, which in 10x Genomics relies on beads with oligos that capture the poly(A) tails on the 3’ ends of mRNA transcripts and adds cell-specific barcodes. To be specific, these barcodes are added with the next GEM beads technology inside the 10x Chromium X apparatus (see the figure). Finally, after cleanup, the produced library undergoes amplification, next-generation sequencing, and data analysis.
10x Genomics GEX Bead

Thousands of oligos outline each 10x bead, containing a 10x barcode unique per bead (i.e., ~ per cell), a unique molecular identifier (UMI), and a template switching oligo that binds at the 3' Poly(A) tail of mature mRNA transcripts.
Source: 10x Genomics
Single Cell ATAC
Single Cell ATAC captures chromatin accessibility events. Accessible, or open, chromatin generally relates to active DNA sites such as those bound by transcription factors, or expressed genes. Hence, ATAC-seq tells you about the epigenetic and transcriptional state of the cells in a sample. This allows you to get a picture of the regulatory elements active in each cell, such as promoters and enhancers.
The technology is based around the Tn5 transposase enzyme. This “tagmentation enzyme” can enter open chromatin, cut DNA, and insert an artificial DNA adapter. The adapters are extended with cell-specific barcodes, which again rely on next GEM beads in the Chromium X. After cleanup, the produced library undergoes amplification, next-generation sequencing, and data analysis.
For a full discussion of the technology, see our blogs on single-cell ATAC-seq:
Combine both techniques with 10x Multiome
Researchers can apply scRNA-seq and scATAC-seq on a sample by dividing it in two and performing both techniques separately on each part. By contrast, 10x Multiome combines the technologies into one experiment. Thus, it can capture transcripts and chromatin accessibility events from each nucleus.
The 10x Multiome protocol starts with nuclei isolation and tagmentation, as in Single Cell ATAC. Then, single-cell barcoding is performed in the Chromium X. However, Single Cell Multiome beads now do the job, which can capture both mRNA poly(A) tails and the adapters added to accessible chromatin regions by Tn5. After cleanup, the two produced libraries undergo separate amplification, collective next-generation sequencing, and data analysis. Consequently, the result is a linked gene expression and chromatin accessibility profile per nucleus.
10x Multiome Bead

Two types of oligos line a 10x Multiome bead. One contains a poly(dT)VN to bind Poly(A) tails of mRNA transcripts, another a spacer that binds the Tn5-adapted DNA fragments of the scATAC library.
Source: 10x Genomics
The 10x Multiome Workflow
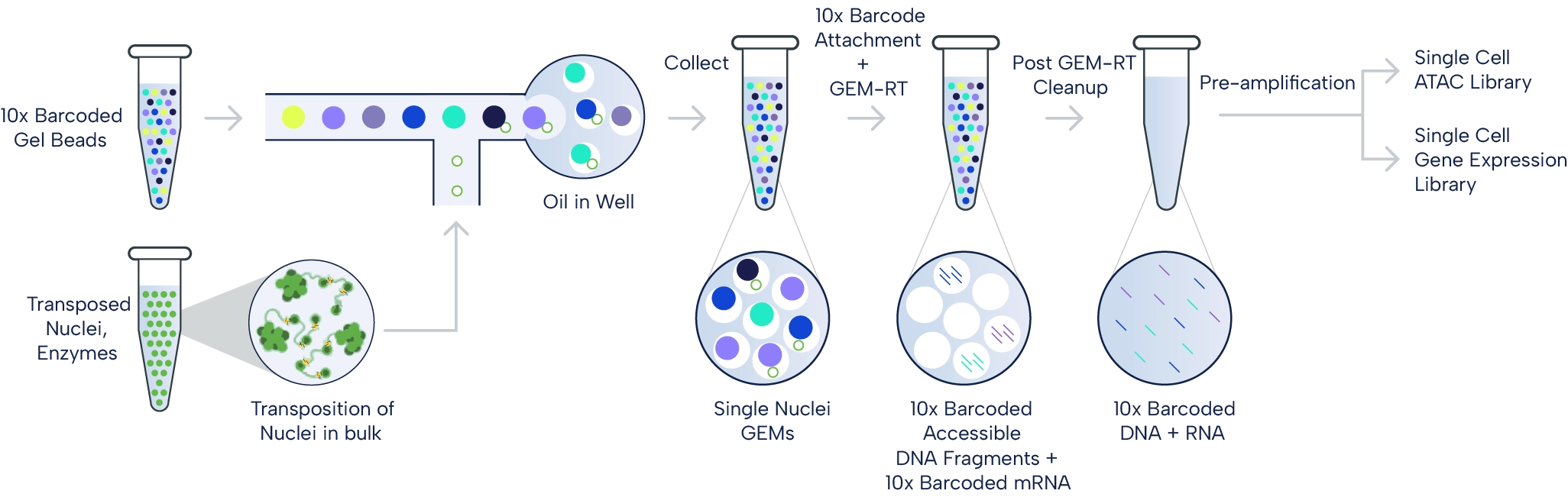
The 10x Multiome workflow.
Nuclei isolation precedes a "tagmentation" step with Tn5 transposase, which cuts open chromatin and tags the DNA fragments with adapters for 10x Barcodes. The nuclei are added to the 10x Chromium X. Here, specialized Next GEM beads capture adapters and mRNAs and add cell-specific and molecule-specific barcodes. Next, the mixture is extracted from the device and pooled. It undergoes library preparation, which includes amplification and cleanup. At Single Cell Discoveries, we perform next-generation sequencing on the Illumina NovaSeq X Plus. Source: 10x Genomics.
Unique benefits of 10x Multiome
With 10x Multiome, we can connect information on the activity of regulatory elements to gene expression data in the same nucleus. That means that there’s a direct one-to-one connection between gene expression and epigenetic programs not capturable otherwise.
On the one hand, from the perspective of a researcher experienced in standalone scATAC-seq, you gain a deep understanding of gene expression dynamics in your sample, validate chromatin accessibility events, and access higher-quality cell type identification.
-
Cross-validate gene expression and chromatin accessibility events
On the other hand, from the perspective of a researcher who is used to standalone scRNA-seq, 10x Multiome offers the additional capacity to identify enhancer and protomer activity. Since enhancers and promoters do not produce mRNA, they are invisible to RNA technology. Yet, the combination with single-cell ATAC-seq enables you to access regulatory network analysis directly on the cell populations for which you generated gene expression profiles.
-
Identify active regulators and map out high-resolution gene networks
What type of opportunity is provided for researchers by 10x Multiome? This, we aim to explain by delving into the specific questions that 10x Multiome is uniquely capable of resolving.
Read the next blog post: 10x Multiome vs. standalone scRNA-seq and scATAC-seq
In the next blog, we cover which questions you can answer with 10x Multiome. In addition, we explain the unique benefits of 10x Multiome and its limitations in comparison to standalone scRNA-seq and scATAC-seq, followed by how you can access 10x Multiome immediately.
More information
Find more information on single-cell ATAC-seq, 10x Genomics, and our approach in our information guide.
Get the 10x Genomics information guide





